Project
Objective: practice a wrap-up project that encompasses most of the workshop
Project - set-up
On your computer, in a folder with a meaningful name, create a new project using the project manager utility on the upper-right part of the rstudio window.
Check if you have all those libraries installed
library("tidyverse")
library("broom")
library("GEOquery")
theme_set(theme_bw(14)) # if you wish to get this theme by defaultAim
As you already experienced, working with GEO datasets can be a hassle. But it provides also a nice exercise as it requires to manipulate of a lot of tables (data.frame and/or matrix). Here, we will investigate the relationship between the expression of ENTPD5 and mir-182 as it was described by Pizzini et al.. Even if the data are already normalised and should be ready to use, you will see that reproducing the claimed results still requires an extensive amount of work.
Retrieve the GEO study
The GEO dataset of interest is GSE35834
- load the study using the
getGEOfunction
Warning
after NCBI moved its pages from http to https make sure that to have GEOquery version > 2.39
- what kind of object is
gse35834?
Tip
To help figuring out the ExpressionSet object, see the figure below. Mind that for this project, the list GSE contains 2 ExpressionSets!
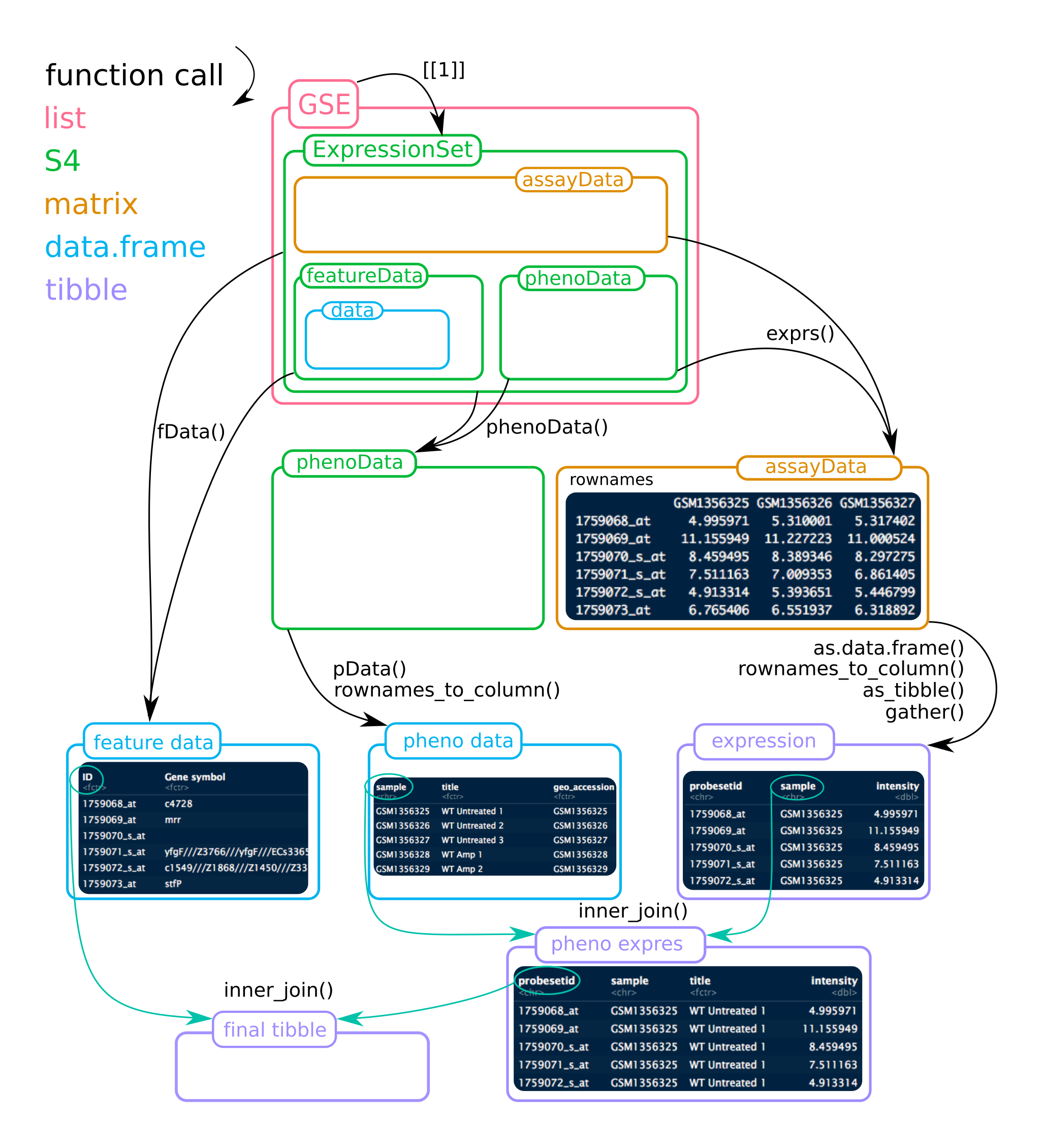
Two platforms were used in this study, which ones?
How can you assign the mRNA or mir data to each element of
gse35834?
Explore the mRNA expression meta-data
You can use phenoData() to get informations on samples or pData() to retrieve them directly as a data.frame.
- Extract the mRNA meta-data as a
tibblewhich you will namerna_meta- rename
geo_accessiontosample - retain only the columns
source_name_ch1and starting with"charact"
- rename
Explore the mir expression meta-data
- Extract the mir meta-data as a
tibblewhich you will namemir_meta- rename
geo_accessiontosample - retain only the columns
source_name_ch1and all starting with “charact”
- rename
Join the meta-data
- Explore the two data frames with
View(rna_meta)andView(mir_meta). Are the samplesGSM*identical?
We would like to somehow join both informations.
Knowing that both data frames have different sample columns, merge them to get the correspondence between RNA GSM* and mir GSM*. Save the result as rna_mir.
Note
If 2 data.frames that are joined (by specific columns) have identical names in their remaining columns, the default suffixes ‘.x’ and ‘.y’ are appended to the concerned column names from the first and second data frames respectively. However, you can make more friendly suffixes that match your actual data using the suffix = c(".x", ".y") option.
Get RNA expression data for the ENTPD5 gene
Expression data can be accessed using exprs() which returns a matrix.
Warning
If you do not pipe the command to head, R would print ALL rows (or until it reaches max.print).
exprs(gse35834[[1]]) %>% head()- rows are probes and columns are sample ids in the form of
GSM*. - Probe ids by themselves are not meaningful, but
fData()provides features.
fData(gse35834[[1]]) %>% head()Again, we need to merge both informations to assign the expression data to the gene of interest.
Find the common values that that allow us to join both data frames.
The
rownamescontain the necessary informations. But as amatrixcontains, by definition, only a single data type (here numerical values), you will need to transform it to adata.frameand convert therownamesto a column usingrownames_to_column(var = "ID").
Save the result asrna_expressionmerge the expression data to the platform annotations (
fData(gse35834[[1]])). Save the result asrna_expression(Don’t worry:Ris always working on temporary objects and you won’t erase the object you are working on).
Note
Warnings about factors being coerced to characters can be ignored.
Find the Entrez gene id for ENTPD5. Usually, the gene symbol is given in the annotation, but each GEO submission is a new discovery.
Filter
rna_expressionfor the gene of interest (ENTPD5) and tidy the samples:
A columnsamplefor allGSM*and a columnrna_expressioncontaining the expression values. Save the result asrna_expression_melt. At this point you should get atibbleof 80 values.Add the meta-data and discard the columns
ID,SPOT_IDandsample_mir. Save the result asrna_expression_melt.
Get mir expression data for miR-182
Repeat the previous step but using
exprs(gse35834[[2]])for themir_expression. This time, the mir names are nicely provided byfData(gse35834[[2]])in the columnmiRNA_ID_LIST.How many rows do you obtain? How many are expected?
Find out what happened, and plot the boxplot distribution of
expressionbyIDFilter out the irrelevant IDs using
greplin thefilterfunction.
Hint
adding ! to a condition means NOT. Example filter(iris, !grepl("a", Species)): remove all Species containing the letter “a”.
- Add the meta-data, count the number of rows. Discard the column
sample_rnaafter joining.
join both expression
Join rna_expression_melt and mir_expression_melt by their common columns EXCEPT sample. Save the result as expression.
Examine gene expression according to meta data
Plot the gene expression distribution by Gender. Is there any obvious difference?
Plot gene AND mir expression distribution by Gender. Is there any obvious difference?
Hint
You will need to tidy by gathering rna and mir expression
Plot gene AND mir expression distributions by source (control / cancer). To make it easier, a quick hack is
separate(expression, source_name_ch1, c("source", "rest"), sep = 12)to getsourceas control / cancer. Is there any difference?Do the same plot as in 3. but reorder the levels so that normal colon appears first. Display normal in “lightgreen” and cancer in “red” using
scale_fill_manual().
plot relation ENTPD5 ~ mir-182 as scatter-plot for all patients
add a linear trend using
geom_smooth()for all data + per sourcedoes it support the claim of the study?
linear models
- get the estimate from the linear trend for each source. linear models are outputted by
lm()as lists. Sincedata.frameare much easier to work with, David Robinson developedbroom.
The estimate of the intercept is not meaningful thus it is filtered out. One can easily see that the slope is not significant when data are slipped by source.
Perform the linear regression and tidy the results for all data, is it significant?
replace
tidybyglanceto extract the \(r^2\). Is this value satisfactory?
Perform a linear model for the expression of ENTPD5 and ALL mirs
Count how many
hsa-mir, which are not star, are present on the array GPL8786Retrieve the expression values for the 677 human mir like you did before. Same procedure, except that you don’t filter for mir-182. Save as
all_mir_rna_expressionPerform the 677 linear models, tidy the results and arrange by the
adj.r.squaredGet the top 12 mir and plot the scatter plot