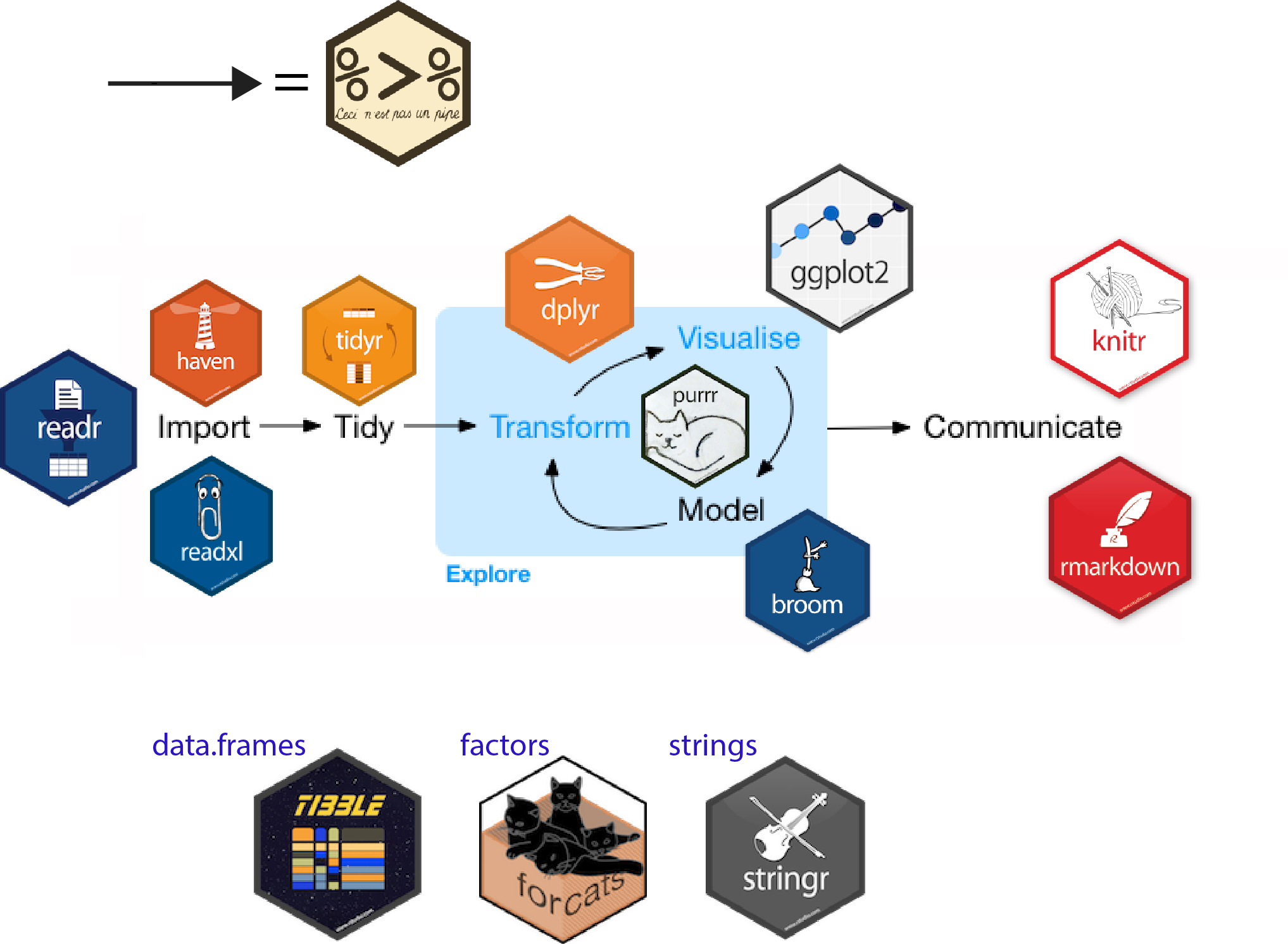
The four day course provides a complete introduction to data science in  with the tidyverse. Focusing on getting data ready, some exploratory analysis, visualization and handling models.
with the tidyverse. Focusing on getting data ready, some exploratory analysis, visualization and handling models.
Preparing data takes up to 90% of the time spent in analysis — speeding this up is the mission of this course
This workshop is composed of 30 hours:
lectures
- ~ 8 hours
- available online
- very short exercises included
- convert them to pdf using chrome
practical sessions
- ~ 15 hours
- using your own laptop
- teachers available
- supplementary exercises if needed
bring your own data
- ~ 7 hours (Day 4)
- teachers available
- project provided for those that don't have data yet