You will learn to:

- use a functional programming approach to simplify your code
- pass functions as arguments to higher order functions
- use
purrr::mapto replaceforloops - deal with nested lists
4 May 2017

purrr::map to replace for loopsEach atomic vector contains only a single type of data
Examples:
# Logical
c(TRUE, FALSE, TRUE)
# double
c(1, 5, 7)
# character
c("character", "sequence")Is the conversion between types
as.*() functionsv_example <- c(1, 3, 7) str(as.character(v_example))
chr [1:3] "1" "3" "7"
v_example <- c(1, 3) str(v_example)
num [1:2] 1 3
str(c(v_example, "seven"))
chr [1:3] "1" "3" "seven"
Adapted from the tutorial of Jenny Bryan
example
my_list <- list(1, 3, "seven") str(my_list)
List of 3 $ : num 1 $ : num 3 $ : chr "seven"
is.vector(my_list)
[1] TRUE
is.atomic(my_list)
[1] FALSE
Adapted from the tutorial of Jenny Bryan

purrr enhances R's functional programming toolkit by providing a complete and consistent set of tools for working with functions and vectors (purrr overview on github page)
functional programming is a programming paradigm – a style of building the structure and elements of computer programs – that treats computation as the evaluation of mathematical functions and avoids changing-state and mutable data Wikipedia
FOR EACH x DO f
Calculate the mean of each column of the mtcars dataset
for loopsmeans <- vector("double", ncol(mtcars))
for (i in seq_along(mtcars)) {
means[i] <- mean(mtcars[[i]])
}
means
[1] 20.090625 6.187500 230.721875 [4] 146.687500 3.596563 3.217250 [7] 17.848750 0.437500 0.406250 [10] 3.687500 2.812500
apply(mtcars, 2, mean) %>% str()
Named num [1:11] 20.09 6.19 230.72 146.69 3.6 ... - attr(*, "names")= chr [1:11] "mpg" "cyl" "disp" "hp" ...
lapply(mtcars, mean) %>% str()
List of 11 $ mpg : num 20.1 $ cyl : num 6.19 $ disp: num 231 $ hp : num 147 $ drat: num 3.6 $ wt : num 3.22 $ qsec: num 17.8 $ vs : num 0.438 $ am : num 0.406 $ gear: num 3.69 $ carb: num 2.81
From R for Data Science
| function | input | output |
|---|---|---|
apply() |
array | vector or array or list |
lapply() |
list | list |
sapply() |
list | vector or array or list |
vapply() |
list | specified but inconsistent |
apply() family are inconsistent.lapply()apply() family still useful to avoid dependencies (package development)purrr::map() family of functionsmap() is the general function and close to base::lapply()map() introduces shortcuts (absent in lapply())map_lgl(), map_int(), map_dbl(), map_chr(), map_df(), walk())Purrr::map()purrr::map() is type stable
map(YOUR_LIST, YOUR_FUNCTION)antennate <- function(x) put_on(x, antenna) map(legos, antennate)

legos

antenna

map(legos, antennate)
Split the mtcars dataset by each value of cylinder
spl_mtcars <- mtcars %>% split(.$cyl) str(spl_mtcars, max.level = 1)
List of 3 $ 4:'data.frame': 11 obs. of 11 variables: $ 6:'data.frame': 7 obs. of 11 variables: $ 8:'data.frame': 14 obs. of 11 variables:
From R for Data Science
purrr::map() reminder:
map(YOUR_LIST, YOUR_FUNCTION) # is equivalent to: YOUR_LIST %>% map(YOUR_FUNCTION)
YOUR_LIST = spl_mtcarsYOUR_FUNCTION can be an anonymous function (declared on the fly)
str(spl_mtcars, max.level = 1)
List of 3 $ 4:'data.frame': 11 obs. of 11 variables: $ 6:'data.frame': 7 obs. of 11 variables: $ 8:'data.frame': 14 obs. of 11 variables:
spl_mtcars %>% map(function(df) lm(mpg ~ wt, data = df))
$`4`
Call:
lm(formula = mpg ~ wt, data = df)
Coefficients:
(Intercept) wt
39.571 -5.647
$`6`
Call:
lm(formula = mpg ~ wt, data = df)
Coefficients:
(Intercept) wt
28.41 -2.78
$`8`
Call:
lm(formula = mpg ~ wt, data = df)
Coefficients:
(Intercept) wt
23.868 -2.192 purrr::map() spl_mtcars %>% map(function(df) lm(mpg ~ wt, data = df)) %>% map(summary) %>% map(function(x) x$r.squared)
$`4` [1] 0.5086326 $`6` [1] 0.4645102 $`8` [1] 0.4229655

The code above can be simplified using shortcuts provided by purrr
purrr::map() One sided formula to create anonymous functions (using ~ and the placeholder . which refers to the current list element)
map(YOUR_LIST, function(df) lm(mpg ~ wt, data = df)) # is equivalent to: map(YOUR_LIST, ~ lm(mpg ~ wt, data = .))
Our previous code can be simplified:
spl_mtcars %>% map(function(df) lm(mpg ~ wt, data = df)) %>% map(summary) %>% map(function(x) x$r.squared)
spl_mtcars %>% map(~lm(mpg ~ wt, data = .)) %>% map(summary) %>% map(~.$r.squared)
spl_mtcars %>%
map(~lm(mpg ~ wt, data = .)) %>%
map(summary) %>%
map("r.squared")purrr::map() (like base base::lapply())
map() makes a list.map_lgl() makes a logical vector.map_int() makes an integer vector.map_dbl() makes a double vector.map_chr() makes a character vector.map_df() makes a data frame (row-binding elements)map()spl_mtcars %>%
map(~lm(mpg ~ wt, data = .)) %>%
map(summary) %>%
map("r.squared") %>%
str()
List of 3 $ 4: num 0.509 $ 6: num 0.465 $ 8: num 0.423
map_dbl()spl_mtcars %>%
map(~lm(mpg ~ wt, data = .)) %>%
map(summary) %>%
map_dbl("r.squared") %>%
str()
Named num [1:3] 0.509 0.465 0.423 - attr(*, "names")= chr [1:3] "4" "6" "8"
deaths MS Excel example file
path using the following command: path <- readxl_example("deaths.xls")readr lecture!library(readxl)
path <- readxl_example("deaths.xls")
path %>%
excel_sheets() %>%
map_df(read_excel, path = path, range = "A5:F15")
# A tibble: 20 x 6
Name Profession Age `Has kids` `Date of birth`
<chr> <chr> <dbl> <lgl> <dttm>
1 David Bowie musician 69 TRUE 1947-01-08
2 Carrie Fisher actor 60 TRUE 1956-10-21
3 Chuck Berry musician 90 TRUE 1926-10-18
4 Bill Paxton actor 61 TRUE 1955-05-17
5 Prince musician 57 TRUE 1958-06-07
6 Alan Rickman actor 69 FALSE 1946-02-21
7 Florence Henderson actor 82 TRUE 1934-02-14
8 Harper Lee author 89 FALSE 1926-04-28
9 Zsa Zsa Gábor actor 99 TRUE 1917-02-06
10 George Michael musician 53 FALSE 1963-06-25
11 Vera Rubin scientist 88 TRUE 1928-07-23
12 Mohamed Ali athlete 74 TRUE 1942-01-17
13 Morley Safer journalist 84 TRUE 1931-11-08
14 Fidel Castro politician 90 TRUE 1926-08-13
15 Antonin Scalia lawyer 79 TRUE 1936-03-11
16 Jo Cox politician 41 TRUE 1974-06-22
17 Janet Reno lawyer 78 FALSE 1938-07-21
18 Gwen Ifill journalist 61 FALSE 1955-09-29
19 John Glenn astronaut 95 TRUE 1921-07-28
20 Pat Summit coach 64 TRUE 1952-06-14
# ... with 1 more variables: `Date of death` <dttm>map_df() arguments.id argumentset_names()to set names in a vector
set_names(1:5)
1 2 3 4 5 1 2 3 4 5
set_names(1:5, nm = letters[1:5])
a b c d e 1 2 3 4 5
is a wrapper for setNames which does some checkings and is stricter
setNames(1:5, nm = letters[1:2])
a b <NA> <NA> <NA> 1 2 3 4 5
set_names(1:5, nm = letters[1:2])
Error: `x` and `nm` must be the same length
path %>% excel_sheets() %>% set_names() %>% # creates a named vector map_df(read_excel, path = path, range = "A5:F15", .id = "profession_area") %>% janitor::clean_names()
# A tibble: 20 x 7
profession_area name profession age has_kids date_of_birth
<chr> <chr> <chr> <dbl> <lgl> <dttm>
1 arts David Bowie musician 69 TRUE 1947-01-08
2 arts Carrie Fisher actor 60 TRUE 1956-10-21
3 arts Chuck Berry musician 90 TRUE 1926-10-18
4 arts Bill Paxton actor 61 TRUE 1955-05-17
5 arts Prince musician 57 TRUE 1958-06-07
6 arts Alan Rickman actor 69 FALSE 1946-02-21
7 arts Florence Henderson actor 82 TRUE 1934-02-14
8 arts Harper Lee author 89 FALSE 1926-04-28
9 arts Zsa Zsa Gábor actor 99 TRUE 1917-02-06
10 arts George Michael musician 53 FALSE 1963-06-25
11 other Vera Rubin scientist 88 TRUE 1928-07-23
12 other Mohamed Ali athlete 74 TRUE 1942-01-17
13 other Morley Safer journalist 84 TRUE 1931-11-08
14 other Fidel Castro politician 90 TRUE 1926-08-13
15 other Antonin Scalia lawyer 79 TRUE 1936-03-11
16 other Jo Cox politician 41 TRUE 1974-06-22
17 other Janet Reno lawyer 78 FALSE 1938-07-21
18 other Gwen Ifill journalist 61 FALSE 1955-09-29
19 other John Glenn astronaut 95 TRUE 1921-07-28
20 other Pat Summit coach 64 TRUE 1952-06-14
# ... with 1 more variables: date_of_death <dttm>map(legos, antennate)


map(legos, antennate)
map2()

enhair <- function(x, y) x %>% put_on(y) map2(legos, hairs, enhair)
dplyr
tibble introduces list-columnsdplyrtidyr::nest()purrr::map
tibble(numbers = 1:8,
my_list = list(a = c("a", "b"), b = 2.56,
c = c("a", "b"), d = rep(TRUE, 4),
d = 2:3, e = 4:6, f = "Z", g = 1:4))
# A tibble: 8 x 2
numbers my_list
<int> <list>
1 1 <chr [2]>
2 2 <dbl [1]>
3 3 <chr [2]>
4 4 <lgl [4]>
5 5 <int [2]>
6 6 <int [3]>
7 7 <chr [1]>
8 8 <int [4]>mtcars %>% group_by(cyl) %>% nest()
# A tibble: 3 x 2
cyl data
<dbl> <list>
1 6 <tibble [7 x 10]>
2 4 <tibble [11 x 10]>
3 8 <tibble [14 x 10]>mtcars %>%
group_by(cyl) %>%
nest() %>%
mutate(model = map(data, ~lm(mpg ~ wt, data = .)),
summary = map(model, summary),
r_squared = map_dbl(summary, "r.squared"))
# A tibble: 3 x 5
cyl data model summary r_squared
<dbl> <list> <list> <list> <dbl>
1 6 <tibble [7 x 10]> <S3: lm> <S3: summary.lm> 0.4645102
2 4 <tibble [11 x 10]> <S3: lm> <S3: summary.lm> 0.5086326
3 8 <tibble [14 x 10]> <S3: lm> <S3: summary.lm> 0.4229655dplyr, tidyr, tibble, purrr and broom nicely work togetherx1 <- list(c(1, 2), c(3, 4)) x2 <- list(list(1, 2), list(3, 4)) x3 <- list(1, list(2, list(3)))
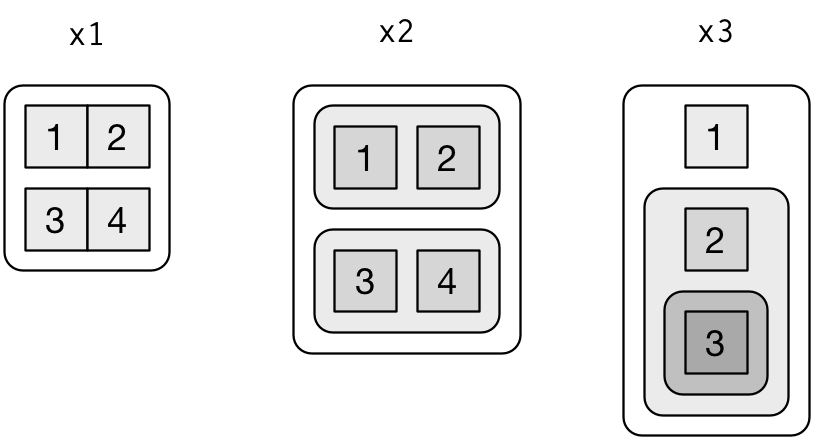
from R for Data Science
flatten() functionflatten()flatten_int()flatten_dbl()flatten_lgl()flatten_chr()x <- list(list(a = 1, b = 2), list(c = 3, d = 4)) flatten(x) %>% str()
List of 4 $ a: num 1 $ b: num 2 $ c: num 3 $ d: num 4
flatten(x) %>% flatten_dbl() %>% str()
num [1:4] 1 2 3 4
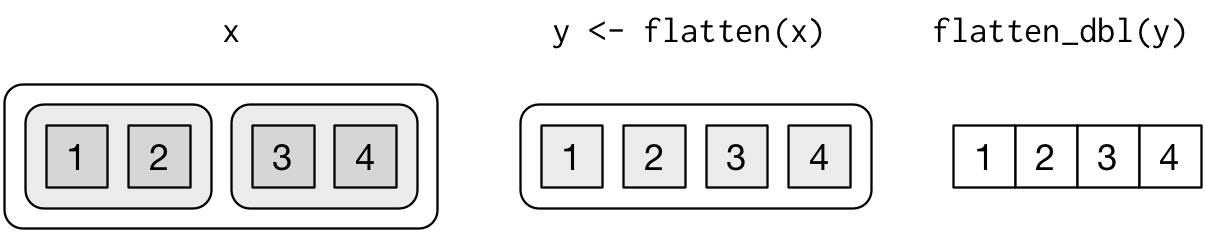
from R for Data Science
base::unlist() vs flatten()unlist() but flatten() is more consistent
unlist() will always succeedunlist(1:3)
[1] 1 2 3
flatten(1:3)
Error: `.x` must be a list (integer)
my_list <- list("One", 2, 3)
unlist(my_list) %>% str()
chr [1:3] "One" "2" "3"
flatten(my_list) %>% str()
List of 3 $ : chr "One" $ : num 2 $ : num 3
flatten_dbl(my_list)
Error: Can't coerce element 1 from a character to a double
x <- list(
x = list(a = 1,
b = 3,
c = 5),
y = list(a = 2,
b = 4,
c = 6)
)
x %>% str()List of 2 $ x:List of 3 ..$ a: num 1 ..$ b: num 3 ..$ c: num 5 $ y:List of 3 ..$ a: num 2 ..$ b: num 4 ..$ c: num 6
x %>% transpose() %>% str()
List of 3 $ a:List of 2 ..$ x: num 1 ..$ y: num 2 $ b:List of 2 ..$ x: num 3 ..$ y: num 4 $ c:List of 2 ..$ x: num 5 ..$ y: num 6
l1 <- list(
group1 = list(
individual1 = list(param1 = 1:2, param2 = 3:4),
individual2 = list(param1 = 5:6, param2 = 7:8)
),
group2 = list(
individual1 = list(param1 = 9:10, param2 = 11:12),
individual2 = list(param1 = 13:14, param2 = 15:16)
)
)
l1 %>% str()List of 2 $ group1:List of 2 ..$ individual1:List of 2 .. ..$ param1: int [1:2] 1 2 .. ..$ param2: int [1:2] 3 4 ..$ individual2:List of 2 .. ..$ param1: int [1:2] 5 6 .. ..$ param2: int [1:2] 7 8 $ group2:List of 2 ..$ individual1:List of 2 .. ..$ param1: int [1:2] 9 10 .. ..$ param2: int [1:2] 11 12 ..$ individual2:List of 2 .. ..$ param1: int [1:2] 13 14 .. ..$ param2: int [1:2] 15 16

listviewer::jsonedit()listviewer packagel1 %>% listviewer::jsonedit()
# Extract individual2 using a string shortcut
l1 %>%
map("individual2") %>% str()
List of 2 $ group1:List of 2 ..$ param1: int [1:2] 5 6 ..$ param2: int [1:2] 7 8 $ group2:List of 2 ..$ param1: int [1:2] 13 14 ..$ param2: int [1:2] 15 16
# chain 2 map calls to get param1 out of both groups
l1 %>%
map("individual2") %>%
map("param1") %>% str()
List of 2 $ group1: int [1:2] 5 6 $ group2: int [1:2] 13 14
map()!l1 %>%
map("individual2") %>%
map("param1") %>% str()
List of 2 $ group1: int [1:2] 5 6 $ group2: int [1:2] 13 14
l1 %>%
map(c("individual2", "param1")) %>% str()
List of 2 $ group1: int [1:2] 5 6 $ group2: int [1:2] 13 14
map()l1 %>% map(~map(., ~map(., mean))) %>% str()
List of 2 $ group1:List of 2 ..$ individual1:List of 2 .. ..$ param1: num 1.5 .. ..$ param2: num 3.5 ..$ individual2:List of 2 .. ..$ param1: num 5.5 .. ..$ param2: num 7.5 $ group2:List of 2 ..$ individual1:List of 2 .. ..$ param1: num 9.5 .. ..$ param2: num 11.5 ..$ individual2:List of 2 .. ..$ param1: num 13.5 .. ..$ param2: num 15.5
at_depth()map()map a function to a lower level of a list
x %>% at_depth(0, fun) # is equivalent to # fun(x) x %>% at_depth(1, fun) # is equivalent to # map(x, fun) x %>% at_depth(2, fun) # is equivalent to # map(x, . %>% map(fun))
l1 %>% map(~map(., ~map(., mean)))
l1 %>% at_depth(3, mean) %>% str()
List of 2 $ group1:List of 2 ..$ individual1:List of 2 .. ..$ param1: num 1.5 .. ..$ param2: num 3.5 ..$ individual2:List of 2 .. ..$ param1: num 5.5 .. ..$ param2: num 7.5 $ group2:List of 2 ..$ individual1:List of 2 .. ..$ param1: num 9.5 .. ..$ param2: num 11.5 ..$ individual2:List of 2 .. ..$ param1: num 13.5 .. ..$ param2: num 15.5
combines a list from the left
1:4 %>% reduce(`-`)
[1] -8
((1 - 2) - 3) - 4
[1] -8
combines a list from the right
1:4 %>% reduce_right(`-`)
[1] -2
1 - (2 - (3 - 4))
[1] -2
1:4 %>% reduce(`+`) c(1, 2, 3, 4) c( 3, 3, 4) c( 6, 4) c( 10)
my_add <- function(x, y) {
print(paste("x =", x, "y =", y))
x + y
}reduce(1:4, my_add)
[1] "x = 1 y = 2" [1] "x = 3 y = 3" [1] "x = 6 y = 4"
[1] 10
reduce_right(1:4, my_add)
[1] "x = 4 y = 3" [1] "x = 7 y = 2" [1] "x = 9 y = 1"
[1] 10
similar to reduce() but keeps intermediate results
1:4 %>% accumulate(`+`)
[1] 1 3 6 10
accumulates a list from the right
1:4 %>% accumulate_right(`+`)
[1] 10 9 7 4
Make your pure #rstats functions purr with purrr, a new package for functional programming: http://t.co/91Efuz0txk
— Hadley Wickham (@hadleywickham) 29 septembre 2015
A function is called "pure" if all its inputs are declared as inputs - none of them are hidden - and likewise all its outputs are declared as outputs Kris Jenkins
start <- 10
impure <- function(x) {
print(start)
x + start
}
result <- impure(2)
[1] 10
result
[1] 12
pure <- function(x, start) {
x + start
}
result <- pure(2, start)
result
[1] 12
log() has side-effectspurrr::safely() to catch every outputlog()(res <- log(10))
[1] 2.302585
res <- log("a")
Error in log("a"): non-numeric argument to mathematical function
res
[1] 2.302585
log()safe_log <- purrr::safely(log) (res <- safe_log(10))
$result [1] 2.302585 $error NULL
res <- safe_log("a")
res
$result NULL $error <simpleError in .f(...): non-numeric argument to mathematical function>
Let's go further with the safe_log() example
# Example from the help page
x <- list("a", 1, 2)
y <- x %>% map(safely(log))
y %>% str()List of 3 $ :List of 2 ..$ result: NULL ..$ error :List of 2 .. ..$ message: chr "non-numeric argument to mathematical function" .. ..$ call : language .f(...) .. ..- attr(*, "class")= chr [1:3] "simpleError" "error" "condition" $ :List of 2 ..$ result: num 0 ..$ error : NULL $ :List of 2 ..$ result: num 0.693 ..$ error : NULL
y %>% transpose() %>% str()
List of 2 $ result:List of 3 ..$ : NULL ..$ : num 0 ..$ : num 0.693 $ error :List of 3 ..$ :List of 2 .. ..$ message: chr "non-numeric argument to mathematical function" .. ..$ call : language .f(...) .. ..- attr(*, "class")= chr [1:3] "simpleError" "error" "condition" ..$ : NULL ..$ : NULL
walkmap() to call functions for its side effects
%>%)x <- list(1, "a", 3) x %>% walk(print)
[1] 1 [1] "a" [1] 3
x %>% walk(print) %>% str()
[1] 1 [1] "a" [1] 3 List of 3 $ : num 1 $ : chr "a" $ : num 3
x %>% map(print) %>% str()
[1] 1 [1] "a" [1] 3 List of 3 $ : num 1 $ : chr "a" $ : num 3
x %>% walk(my_print) %>% str()
[1] 1 [1] "a" [1] 3 List of 3 $ : num 1 $ : chr "a" $ : num 3
x %>% map(my_print) %>% str()
[1] 1 [1] "a" [1] 3 List of 3 $ : NULL $ : NULL $ : NULL
