Definitions
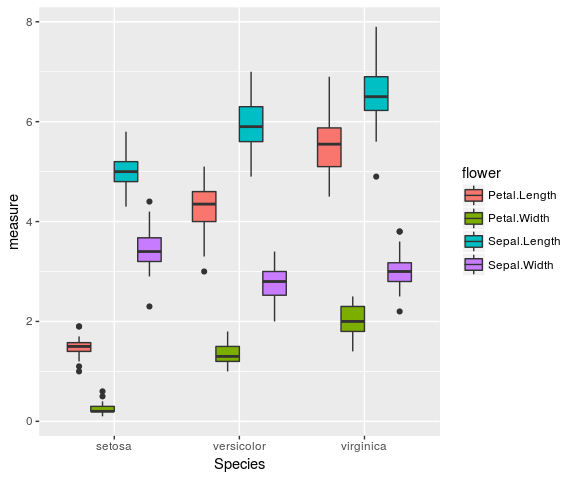
- Principles of tidy data to structure data
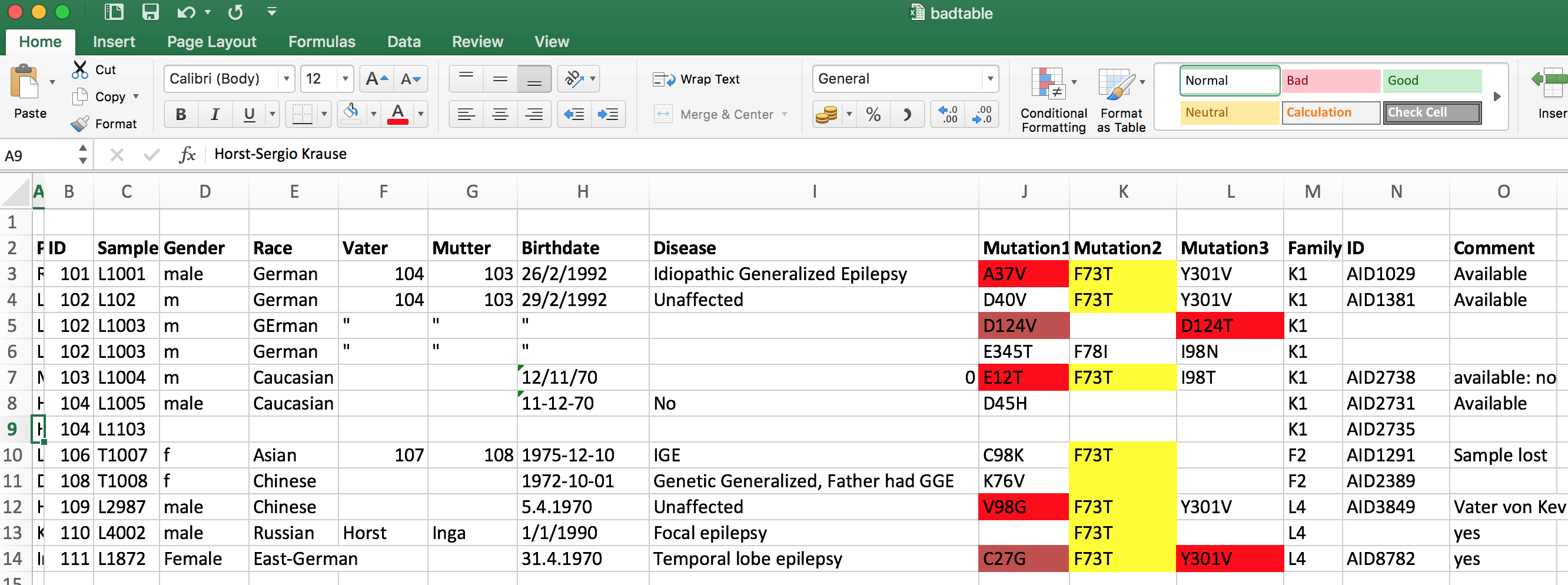
- Find errors in existing data sets
- Structure data
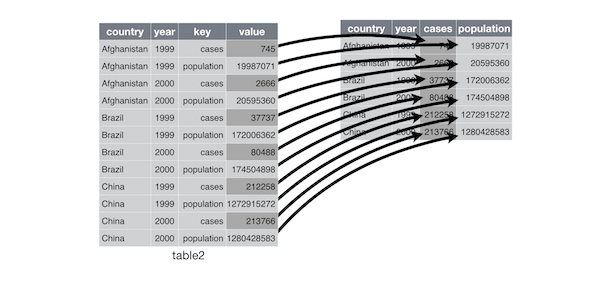
- Reshaping data with tidyr
Comments
- Cleaning data also requires
dplyr tidyranddplyrare intertwined- Focus on "tidy data"
- Introduction of
tidyrways