3rd June 2016
R workshop
Day 2 - advanced
Modern R
David Robinson summarized the goal on his laptop
see also what Karl Broman is recommanding for people who learnt R a while ago
Modern pipeline
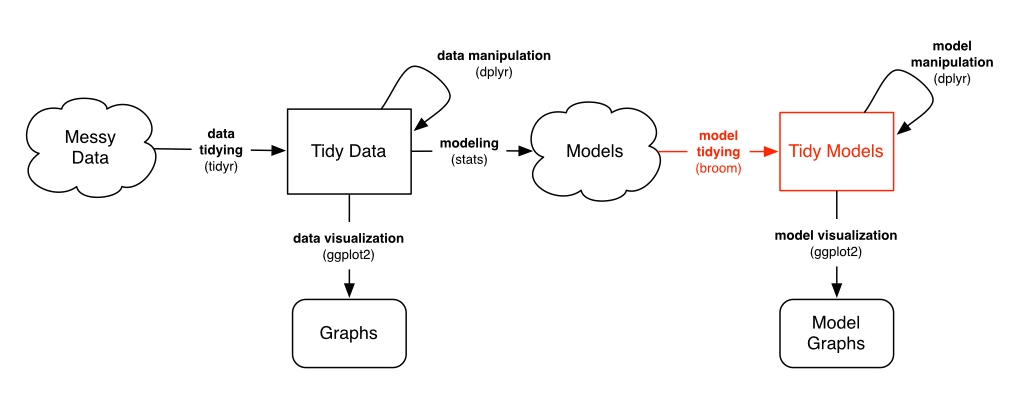
source: David Robinson check out David's broom presentation
reading data
Hadley Wickham and Wes McKinney recently released feather, a new python / R project.
It rapidly stores dataframes as binary files and preserves column types.
Managing multiple models
Tutorial based on the great conference by Hadley Wickham
purrr::map / dplyr::do
progress bar will be added

Functional programming and nested data_frames
Using purrr and tidyr. Hadley is focusing on every part of R to clean it up.
purrr revisits the apply family in a consistent way. tidyr::nest nests list in tibble::data_frame to keep related things together.
For loops emphasise on objects and not actions
compare (notice seq_along instead of 1:length(mtcars))
means <- vector("double", ncol(mtcars))
for (i in seq_along(mtcars)) {
means[i] <- mean(mtcars[[i]])
}
means
## [1] 20.090625 6.187500 230.721875 146.687500 3.596563 3.217250 ## [7] 17.848750 0.437500 0.406250 3.687500 2.812500
and
library("purrr")
map_dbl(mtcars, mean)
## mpg cyl disp hp drat wt ## 20.090625 6.187500 230.721875 146.687500 3.596563 3.217250 ## qsec vs am gear carb ## 17.848750 0.437500 0.406250 3.687500 2.812500
Nested map
library("purrr")
library("dplyr", warn.conflicts = FALSE)
funs <- list(mean = mean, median = median, sd = sd)
funs %>%
map(~ mtcars %>% map_dbl(.x))
## $mean ## mpg cyl disp hp drat wt ## 20.090625 6.187500 230.721875 146.687500 3.596563 3.217250 ## qsec vs am gear carb ## 17.848750 0.437500 0.406250 3.687500 2.812500 ## ## $median ## mpg cyl disp hp drat wt qsec vs am ## 19.200 6.000 196.300 123.000 3.695 3.325 17.710 0.000 0.000 ## gear carb ## 4.000 2.000 ## ## $sd ## mpg cyl disp hp drat wt ## 6.0269481 1.7859216 123.9386938 68.5628685 0.5346787 0.9784574 ## qsec vs am gear carb ## 1.7869432 0.5040161 0.4989909 0.7378041 1.6152000
Keep related things together
Linear model per country
library("gapminder")
library("tidyr")
by_country_lm <- gapminder %>%
mutate(year1950 = year - 1950) %>%
group_by(continent, country) %>%
nest() %>%
mutate(model = map(data, ~ lm(lifeExp ~ year1950, data = .x)))
by_country_lm
## Source: local data frame [142 x 4] ## ## continent country data model ## (fctr) (fctr) (chr) (chr) ## 1 Asia Afghanistan <tbl_df [12,5]> <S3:lm> ## 2 Europe Albania <tbl_df [12,5]> <S3:lm> ## 3 Africa Algeria <tbl_df [12,5]> <S3:lm> ## 4 Africa Angola <tbl_df [12,5]> <S3:lm> ## 5 Americas Argentina <tbl_df [12,5]> <S3:lm> ## 6 Oceania Australia <tbl_df [12,5]> <S3:lm> ## 7 Europe Austria <tbl_df [12,5]> <S3:lm> ## 8 Asia Bahrain <tbl_df [12,5]> <S3:lm> ## 9 Asia Bangladesh <tbl_df [12,5]> <S3:lm> ## 10 Europe Belgium <tbl_df [12,5]> <S3:lm> ## .. ... ... ... ...
broom cleanup
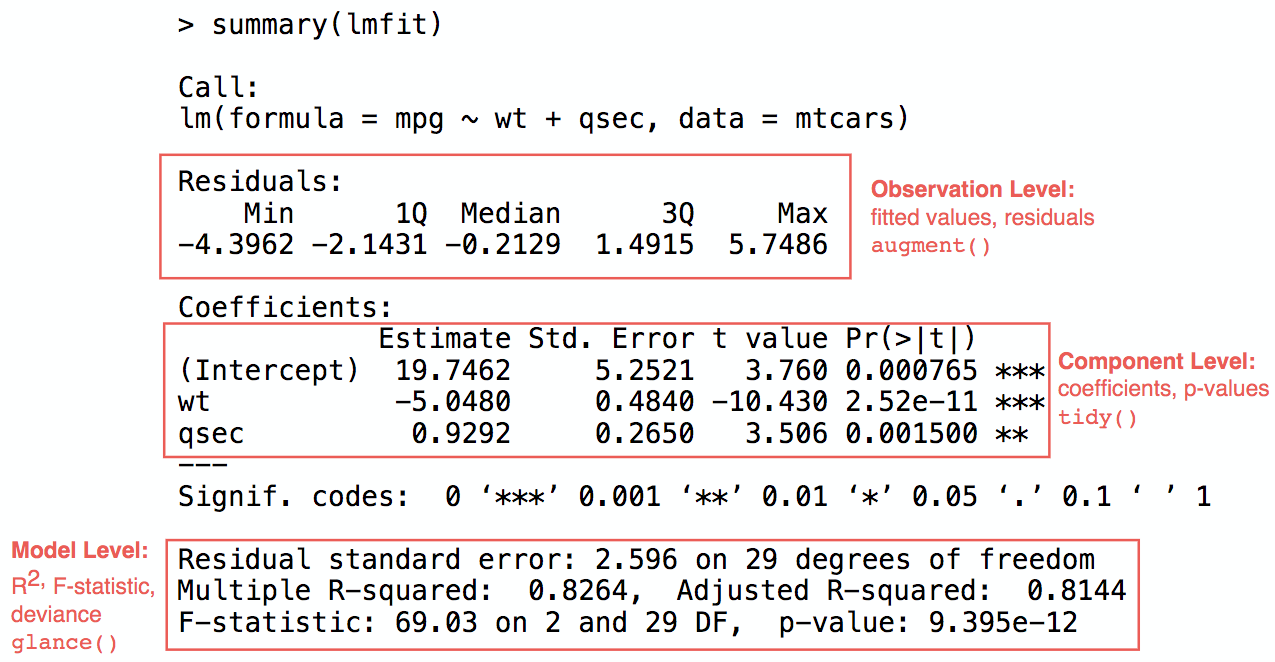
Tidying model coefficients
Use broom to extract, as neat data frames out of lm():
- coefficients estimates: slope and intercept
- \(r^2\)
- residuals
library("broom")
models <- by_country_lm %>%
mutate(glance = map(model, glance),
rsq = glance %>% map_dbl("r.squared"),
tidy = map(model, tidy),
augment = map(model, augment))
models
## Source: local data frame [142 x 8] ## ## continent country data model glance ## (fctr) (fctr) (chr) (chr) (chr) ## 1 Asia Afghanistan <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## 2 Europe Albania <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## 3 Africa Algeria <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## 4 Africa Angola <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## 5 Americas Argentina <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## 6 Oceania Australia <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## 7 Europe Austria <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## 8 Asia Bahrain <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## 9 Asia Bangladesh <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## 10 Europe Belgium <tbl_df [12,5]> <S3:lm> <data.frame [1,11]> ## .. ... ... ... ... ... ## Variables not shown: rsq (dbl), tidy (chr), augment (chr)
Exploratory plots
Does linear models fit all countries?
library("ggplot2")
theme_set(theme_bw(14))
models %>%
ggplot(aes(x = rsq, y = reorder(country, rsq)))+
geom_point(aes(colour = continent))+
theme(axis.text.y = element_text(size = 6))
focus on non-linear trends
models %>% filter(rsq < 0.55) %>% unnest(data) %>% ggplot(aes(x = year, y = lifeExp))+ geom_line(aes(colour = continent))+ facet_wrap(~ country)
shiny - code
library("shiny")
inputPanel(
selectInput("country", "Select Country", levels(models$country))
)
output$rsq <- renderPlot({
models %>%
filter(country == input$country) %>%
unnest(data) %>%
ggplot(aes(x = year, y = lifeExp))+
geom_line(aes(colour = continent))
})
renderUI({
plotOutput("rsq", height = "400", width = "600")
})
shiny
library("shiny")
inputPanel(
selectInput("country", "Select Country", levels(models$country))
)
output$country <- renderPlot({
models %>%
filter(country == input$country) %>%
unnest(data) %>%
ggplot(aes(x = year, y = lifeExp))+
geom_line(aes(colour = continent))
})
renderUI({
plotOutput("country", height = "400", width = "600")
})
shiny - rsquare
library("shiny")
inputPanel(
sliderInput("rsq", "Select rsquared", min = 0, max = 1,
value = c(0, 0.2), dragRange = TRUE)
)
output$rsq <- renderPlot({
models %>%
filter(rsq >= input$rsq[1], rsq <= input$rsq[2]) %>%
unnest(data) %>%
ggplot(aes(x = year, y = lifeExp))+
geom_line(aes(colour = continent))+
facet_wrap(~ country)
})
renderUI({
plotOutput("rsq", height = "400", width = "600")
})
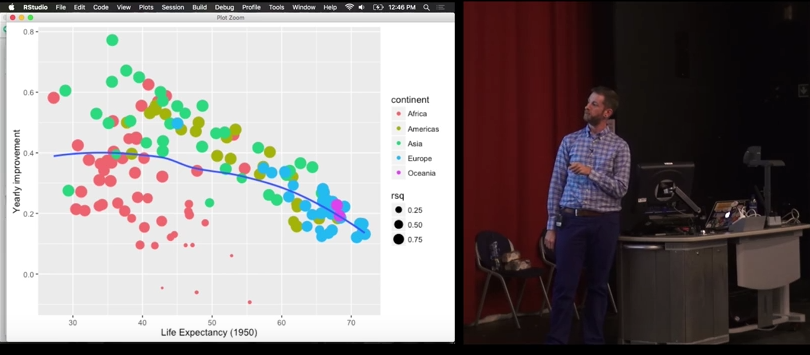
All in all
models %>%
unnest(tidy) %>%
select(continent, country, rsq, term, estimate) %>%
#filter(continent != "Africa") %>%
spread(term, estimate) %>%
ggplot(aes(x = `(Intercept)`, y = year1950))+
geom_point(aes(colour = continent, size = rsq))+
geom_smooth(se = FALSE)+
scale_size_area()+
labs(x = "Life expectancy (1950)",
y = "Yearly improvement")
Error handling
purrr proposes safely() and possibly() to enable error-handling.
safely() is a type-stable version of try. It always returns a list of two elements, the result and the error, and one will always be NULL.
safely(log)(10)
## $result ## [1] 2.302585 ## ## $error ## NULL
safely(log)("a")
## $result ## NULL ## ## $error ## <simpleError in .f(...): non-numeric argument to mathematical function>
to be investigated
Recommended reading
- purrr applied by Ian Lyttle
- R for data science by Hadley
- iterations - purrr by Hadley
- purrr 0.1 by Hadley
- purrr 0.2 by Hadley
Acknowledgments
- Hadley Wickham
- Robert Rudis
- Ian Lyttle
- David Robinson
- Eric Koncina